One of the key components of our National Research Resource Grant is the development of physiologically significant databases. Our data are publicly accessible, in order to promote search in medical image processing and analysis.
Diffusion Tensor Imaging Datasets
Developed by the Laboratory of Brain Anatomical MRI, these datasets show DTI data and three-dimensional fiber trajectories. They feature Raw or Analyze format, with resolution varying from 1.00mm3 to 2.5mm3, and the compressed size varies from 2Mb (for one scan) to 10+Gb (for multiple sessions).
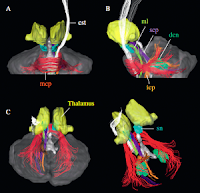
- White Matter Atlas: DTI Atlas of the Brain's White Matter Tracts
This atlas, created by scientists at UCL in Belgium and the Kirby Center in Baltimore, MD, is an online database of fiber tracts. - Human Brain Atlas
These atlases were developed from coronal, sagittal, and axial slices. The CMRM also has a human atlas template, a mouse atlas, a monkey atlas, and a CT-MRI atlas. - DTI Database
This in vivo human database, acquired under the Human Brain Project and the National Research Resource Center grant, contains raw and processed DTI data of the normal population. It is a public database, open to registered users. Its purpose is to facilitate research in DTI data processing and analysis, to study specific biological interests, or to be used as control data. Basic imaging parameters can also be downloaded. Detailed parameters are available upon request. - Developmental DTI Database
These databases showcase pediatric and neonatal DTI datasets. (Coming soon!)
Anatomy Datasets
Developed by the Center for Imaging Science at Johns Hopkins University, these datasets show analyzed biological images and can be used for validating segmentation. These segmented subvolumes consist of Analyze or MATLAB data types. The resolution varies from 0.5mm3 to 1.0mm3. And the compressed size varies from 4.3Mb to 26Mb.
- CAportal: The Computational Anatomy Portal: The Computational Anatomy Portal provides access to the Atlases, Pipelines, Projects and Tools developed at the Johns Hopkins University Center for Imaging Science. Links are also provided to Computational Anatomy Processing Portals.
- CIS Datasets
These biological images have registered several subvolumes, which can be used for segmentation.
Kirby 21: Multi-Modal Resource Reproducibility Study
This project, which showcases many state-of-the-art sequences at the Kirby Center, was spearheaded by Dr. Bennett Landman, while he was a Research Professor at Johns Hopkins University. He now runs the Medical-image Analysis and Statistical Interpretation Lab at Vanderbilt University.
This database consists of acquired scan-rescan imaging sessions on 21 healthy volunteers (no history of neurological disease) at a field of 3 Tesla. Imaging modalities include MPRAGE, FLAIR, DTI, resting state fMRI, B0 and B1 field maps, ASL, VASO, quantitative T1 mapping, quantitative T2 mapping, and magnetization transfer imaging. All data have been converted to NIFTI format. This database is intended to be a resource for statisticians and imaging scientists to be able to quantify the reproducibility of their imaging methods using data available from a generic "1 hour" session at 3T. This effort provides the neuroimaging community with a data resource that may be used for development, optimization, and evaluation of algorithms that exploit the diversity of modern MRI modalities.
The data may contribute to or serve as a priori information for study planning, effect size estimation or as normative reference data from volunteers. Alternatively, the data may be used within an education or tutorial setting and hence provide realistic examples of the data available within clinical research. This database is publicly shared through the "multimodal" project on the NITRC portal (http://www.nitrc.org/projects/multimodal/). The community is encouraged to use this data for investigation of the topics relevant to their research. Investigators who accept the BIRN Repository Data License may access, redistribute, and publish the data without justification of intended use. Forums are provided for researchers who, at their discretion, may seek to coordinate or collaborate on investigations.
The data type consists of NIfTI format, with resolution depending on the image type, and the total compressed size is 8.8Gb for 42 sessions.
Reference: Landman BA, Huang AJ, Gifford A, Vikram DS, Lim IA, Farrell JA, Bogovic JA, Hua J, Chen M, Jarso S, Smith SA, Joel S, Mori S, Pekar JJ, Barker PB, Prince JL, van Zijl PC. Multi-parametric neuroimaging reproducibility: A 3-T resource study. Neuroimage. 2010.
- NITRC: Multi-Modal MRI Reproducibility Resource: Kirby 21
Imaging modalities include MPRAGE, FLAIR, DTI, resting state fMRI, B0 and B1 field maps, ASL, VASO, quantitative T1 mapping, quantitative T2 mapping, and magnetization transfer imaging.
Single-subject Resting state fMRI Reproducibility Resource
A longitudinal single-subject dataset of a healthy male volunteer, 40 years of age at time of initial scan. A total of 158 sessions of MRI data was acquired on a weekly basis, over a span of 185 weeks (a little over 3 years). This is intended to be a resource for statisticians and imaging scientists to be able to quantify the intra-subject inter-session reproducibility of their image analysis methods using data available from a 7 min session at 3T.
- NITRC: Single-subject Resting State fMRI Reproducibility Resource
- 3T Philips Achieva, gradient-echo EPI, 2 s TR, 30 ms TE, SENSE factor 2, 75° flip angle, 37 axial slices, 3x3x3 mm3, 1 mm gap, 16 channel neuro-vascular coil, 200 dynamics (frames), 6:40 scan time, eyes closed during scan.
