A persistent challenge in neuroimaging is the integration of multi-contrast imaging data across micro- , meso-, and macro-scales. Researchers—within our TRDs, Collaborative Projects, and the broader neuroimaging community—routinely work with diverse data types, including volumetric, connectivity, physiological, metabolic, molecular, susceptibility, magnetization transfer, diffusion, and CEST imaging. However, variations in resolution and volume size introduce additional complexity, particularly in aligning data to a common coordinate space using atlases. We focus on developing methods to integrate multimodal, multiscale imaging data to extract latent features that are distributed across domains. Efficient combination of these features can reveal novel biomarkers and accelerate neuroscientific discovery.
We have already demonstrated success in integrating molecular and anatomical data to study diseases such as Alzheimer’s1. Using our multimodal pipeline—combining MRI, spatial transcriptomics2, and clinical metadata—we developed tools for longitudinal atrophy estimation using diffeomorphic mapping and validated them in large, multicenter cohorts. We also created a computational framework to align transcriptomics data with MRI atlases3, addressing challenges from resolution mismatch and anatomical variability. This approach enabled us to link atrophy maps with tau-related gene expression4 and refine spatial characterization of molecular pathology in other datasets5,6. We are extending our success in co-registering MRI with post-mortem histology in preclinical dementia to the alignment of low-resolution clinical scans with high-resolution atlases. This is essential for reusing underutilized clinical data and for creating tools that translate easily to real-world clinical research, ultimately maximizing practical impact.
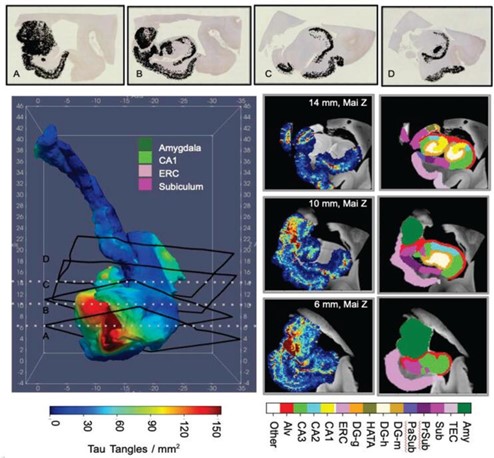
Top: Histology sections with detected Neurofibrillary tangle (NFTs, black) and their 3D locations. Bottom left: Smoothed NFT density over hippocampal subregions using Laplace-Beltrami. Bottom right: 3D NFT slices and matching MRI with manual segmentations at 12 and 18mm (Mai atlas z-axis)8
- Stouffer, K. M. et al. Cross-modality mapping using image varifolds to align tissue-scale atlases to molecular-scale measures with application to 2d brain sections. Nat. communications 15, 3530 (2024).
- Clifton, K. et al. Stalign: Alignment of spatial transcriptomics data using diffeomorphic metric mapping. Nat. communications 14, 8123 (2023).
- Stouffer, K. M., Witter, M. P., Tward, D. J. & Miller, M. I. Projective diffeomorphic mapping of molecular digital pathology with tissue mri. Commun. engineering 1, 44 (2022).
- Stouffer, K. M. et al. Early amygdala and erc atrophy linked to 3d reconstruction of rostral neurofib- rillary tau tangle pathology in alzheimer’s disease. NeuroImage: Clin. 38, 103374 (2023).
- Miller, M., Tward, D. & Trouvé, A. Molecular computational anatomy: Unifying the particle to tissue continuum via measure representations of the brain. BME Front. 2022 (2022).
- Miller, M. I., Trouvé, A. & Younes, L. Space-feature measures on meshes for mapping spatial transcriptomics. Med. Image Analysis 93, 103068 (2024).
